ARVYS Proteins Inc. provides a full spectrum of protein biochemistry services – recombinant protein expression in bacterial, insect and mammalian cells, protein purification, refolding, assays and assay development, protein characterization, fermentation and endotoxin removal.
Outsource your protein biochemistry projects to ARVYS and enjoy superior results, team expertise and customer support after project completion.
Today we look at the publication from Humana Press. Edited by Laure Journet and Eric Cascales Institut de Microbiologie de la Méditerranée, Aix-Marseille Univ – CNRS, Marseille, Franc
Preface
In their ecological niches, bacteria are in contact with other prokaryotic and eukaryotic cells. Bacteria therefore evolved mechanisms to communicate and collaborate with these cells. They also developed belligerent behaviours to eliminate competitors and to infect eukaryotic host cells. These aggressive actions are mediated by effector toxins with specific activities which will ultimately cause target cell lysis or re-routing of metabolic or trafficking pathways in the host. The proper delivery of bacterial effectors into the milieu or directly into target cells is assured by dedicated structures called secretion systems. Up to now, nine secretion systems have been described in bacteria, and additional systems allow the exposition of toxins or adhesins at the cell surface or at the extremity of a pilus structure. These multi-protein trans-envelope complexes differ in composition, mechanism of assembly, and mode of recruitment and transport of toxins. However, studying these macromolecular complexes requires common techniques, ranging from the bioinformatic identification of machine components and effectors to methods of defining interactions amongst the different subunits and the development of reporters to follow effector translocation in vivo. Finally, state-of-the-art techniques have made significant progress in the analysis of these large complexes directly in the bacterial cell or from purified samples. The purpose of this book on bacterial secretion systems is to provide protocols that cover the broad arsenal of techniques used to study a secretion system from A to Z: identifying and localising the different subunits, defining interactions within subunits, monitoring conformational changes, purifying and imaging large complexes, defining the assembly pathway by fluorescence microscopy and the role of energy during assembly or secretion, and identifying secreted effectors as well as using reporters to follow effector transport. Most of these techniques are not restricted to the study of secretion systems but are also of specific interest for any researcher interested in multi-protein complexes of the bacterial cell envelope. The book starts with a chapter describing a recently developed software program aimed at identifying gene clusters encoding secretion systems within bacterial genomes. Then six chapters describe methods for defining the subcellular localisation of the different subunits of a multi-protein system: prediction programs, fractionation, cell surface exposition and isopycnic density gradients to partition inner and outer membranes. Three techniques to determine the topology of membrane proteins (substituted cysteine accessibility, protease accessibility and reporter fusions) are then detailed. This is followed by eleven chapters covering genetic, cytologic and biochemical methods used to study protein–protein and protein–peptidoglycan interactions. Two chapters are dedicated to methods for unveiling the role of energy and conformational changes, and then six chapters describe techniques for purifying and imaging large complexes. Finally, methods to identify effectors as well as the reporter fusions used to validate effector secretion and translocation are presented.
Abstract
Protein secretion systems are complex molecular machineries that translocate proteins through the outer membrane, and sometimes through multiple other barriers. They have evolved by co-option of components from other envelope-associated cellular machineries, making them sometimes difficult to identify and discriminate. Here, we describe how to identify protein secretion systems in bacterial genomes using MacSyFinder. This flexible computational tool uses the knowledge stemming from experimental studies to identify homologous systems in genome data. It can be used with a set of predefined models—“TXSScan”— to identify all major secretion systems of diderm bacteria (i.e., with inner and with LPS-containing outer membranes). For this, it identifies and clusters colocalized components of secretion systems using sequence similarity searches with hidden Markov model protein profiles. Finally, it checks whether the genetic content and organization of clusters satisfy the constraints of the model. TXSScan models can be customized to search for variants of known systems. The models can also be built from scratch to identify novel systems. In this chapter, we describe a complete pipeline of analysis, including the identification of a reference set of experimentally studied systems, the identification of components and the construction of their protein profiles, the definition of the models, their optimization, and, finally, their use as tools to search genomic data.
Areas that stand to be mentioned:
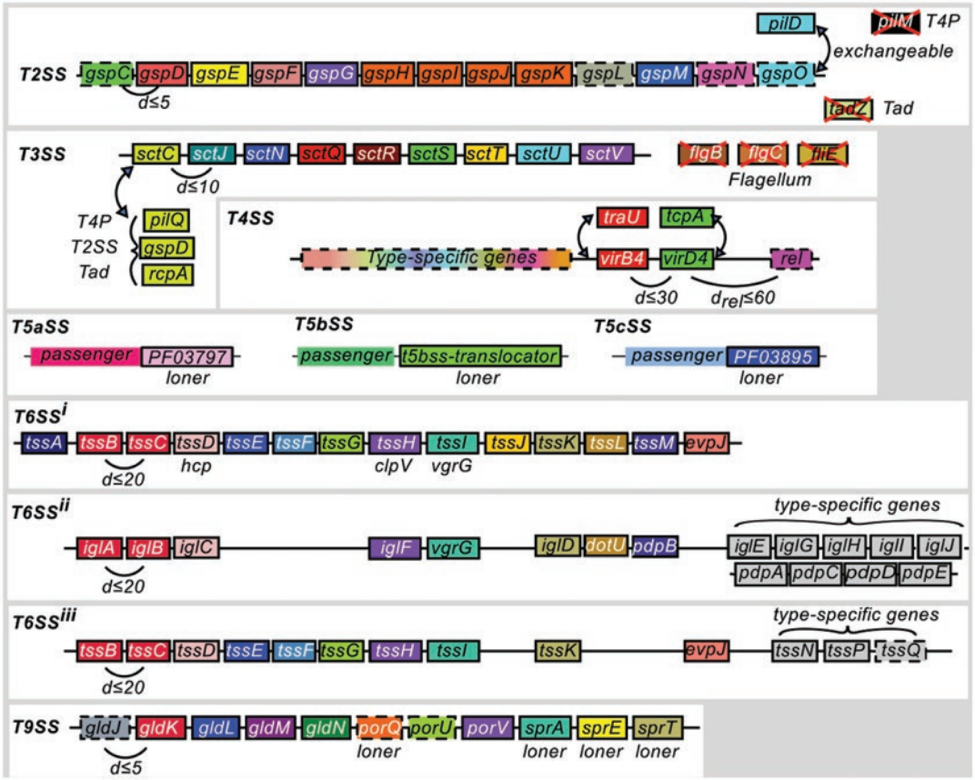
Fig. 1 Models of protein secretion systems available in TXSScan. The T1SS model is displayed in Fig. 2. Each box represents a component and its status in the system’s model: “mandatory” (plain), “accessory” (dashed), or “forbidden” (red cross). Within a system panel: the families of homologous proteins are represented in columns and colored identically. When applicable, the system of origin is indicated next to the box(es). The colocalization parameter of the system is indicated (d). When this parameter is specific to a gene (e.g., the relaxase in T4SS), it is indicated by a subscript (e.g., drel). Additional features specific to a component can be found in Table 2. Curved double-headed arrows indicate exchangeable components. Figures and legends are freely reproduced with modification from reference [9] [as specified by the Creative Commons Attribution (CC BY) license version 4.0]
To buy the full journal, click here.
ARVYS Proteins Inc. provides a full spectrum of protein services to the life science, pharmaceutical and biotechnology communities. Our work experience encompasses almost every aspect of protein biochemistry allowing us to contribute to projects regardless of whether they are at early research or late development stages. We can be your partner in:
– generation and expression of recombinant proteins in bacterial, baculovirus and mammalian expression systems,
– large-scale fermentation,
– cell culture,
– purification of recombinant proteins, antibodies or naturally occurring proteins,
– refolding from inclusion bodies,
– improvement of protein stability,
– protein labeling with fluorescent, biotin and enzyme probes,
– endotoxin removal and testing for in vivo studies,
– protein characterization to monitor its integrity and functionality
“Don’t think clean thoughts with dirty enzymes” – is a famous saying of Efraim Racker, one of the founding fathers of modern biochemistry. We believe this philosophy should be extended to all proteins – purified and well-characterized preparations will save you time, effort and resources. Your confidence in protein reagents is the core focus of our business.
ARVYS Proteins Inc. is a Contract Research Organization (CRO) that Specializes in Custom Protein Services for Drug Discovery and Life Science Research.