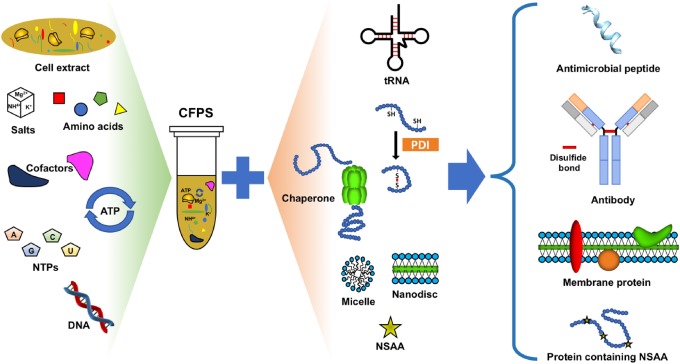
Overview
The ability to synthesize proteins using recombinant DNA techniques has revolutionized molecular and cell biology research by enabling the study of target proteins in a laboratory setting. Most commonly, this is done by transforming live cells with a plasmid that contains the gene for the protein of interest and transcriptional activators. The affordability and high yields this method produces make it a desirable first choice for those seeking to study a particular protein of interest. However, in vivo protein expression is very laborious and time-consuming, often taking days to weeks to complete a single preparation. In recent years, several methods concerning in vitro protein expression have been developed that reduce this multi-day approach to a single day, sometimes even a few hours with the right amount of preparation. Cell-free protein synthesis (CFPS), also commonly referred to as in-vitro translation (IVT), seeks to enable researchers to produce proteins in the lab without spending multiple days or weeks culturing cells. While many CFPS systems have been characterized (E. coli cell lysates, wheat germ extracts, etc.) the process is generally the same: cells are cultured and lysed to produce a whole-cell extract that contains all of the biological machinery necessary to synthesize a protein from a gene sequence (DNA/RNA polymerases, ribosomes, chaperones, etc.). This extract can then be added to a master mix that contains all of the other necessary components, namely those necessary for the synthesis of all proteins (all essential amino acids, ATP/GTP energy sources for powering transcriptional/translational enzymes). Ancillary components can also be added to this master mix that is specific for certain proteins of interest, such as cofactors for metal-binding proteins or isotopic amino acids for synthesizing radiolabeled proteins. Once the final reaction mix is assembled, genetic information in the form of an mRNA sequence or even a recombinantly produced plasmid/linear expression template can be added to generate the protein of interest. CFPS is fast, easy, and has several advantages/disadvantages over traditional in vivo protein synthesis.
Advantages
As mentioned previously, the main advantage of CFPS is its speed. All components necessary for this reaction can be prepared in advance (cell extract, reaction master mix, genetic information) and stored for long periods. Rather than scheduling days of work to culture cells for expression, running a CFPS experiment is as trivial as thawing all of these components, setting up the reactions, and letting them run. Despite enabling this highly convenient workflow, the open environment of CFPS provides additional benefits. First, the lack of an enclosed environment means that reactions can be monitored in real-time using spectrophotometry. Samples for gel electrophoresis can be taken at various time points throughout the synthesis reaction to test for the presence of properly produced protein. CFPS is also commonly used for synthesizing proteins that are toxic to cells, and thus cannot be made in large quantities from the traditional in vivo method. These toxic proteins rarely affect the transcription/translation machinery necessary for producing them, thus CFPS is not negatively impacted by their presence. Lastly, CFPS allows for the easy incorporation of unnatural or modified amino acids into protein structure. These amino acids can simply be added to the master mix to ensure their eventual incorporation into the finished protein. This technique is ideal for producing radiolabeled protein, wherein amino acids that contain radioactive isotopes are added to the master mix so that the final protein can be quantified using a scintillation counter.
Limitations
Even though there are many merits to CFPS, it has a few major flaws that prevent it from being the de-facto technique for laboratory protein synthesis, the first being the overall yield. The typical protocols for CFPS are simply incapable of producing large quantities of protein that are required for structural techniques or proteomics. While it is possible to scale up CFPS, the materials required to satisfy the high concentrations required for those techniques are simply not feasible under typical laboratory conditions, especially when compared to in vivo protein expression. Due to this limitation, CFPS is most commonly used for applications where only small quantities of a protein of interest are required. The second major limitation is sensitivity to nucleases or proteases. CFPS relies on the uninterrupted pairing of transcription and translation, meaning that mRNA messengers and final protein products are highly susceptible to degradation by nucleases/proteases, respectively, that are a part of the cell extract. While this is also an issue with in vivo protein synthesis, it is especially a problem for CFPS because of the lack of functional compartmentalization between individual cellular components. In other words, the cell extract contains the nucleases/proteases of many different cells whereas in vivo synthesis would contain discrete populations of protein products and enzymes.
Should I Use Cell-Free Protein Synthesis?
The main consideration in whether a researcher should use CFPS is the desired application for the protein that is being produced. Is it one where a large concentration per sample is necessary? Does this application require multiple trials with the same protein? If the answer to either of these questions is “yes”, then CFPS is not the best expression choice. However, if low concentration samples will suffice, and many different proteins will need to be tested as part of this experiment, then CFPS will save countless hours of lab time due to its speed and compatibility with high-throughput experimentation. If none of these criteria apply and the experiment simply requires a small amount of protein that is toxic to cells or contains unnatural amino acids, then CFPS may also be the best choice for you.